NucMM Challenge: Neuronal Nuclei Segmentation at Sub-Cubic Millimeter Scale¶
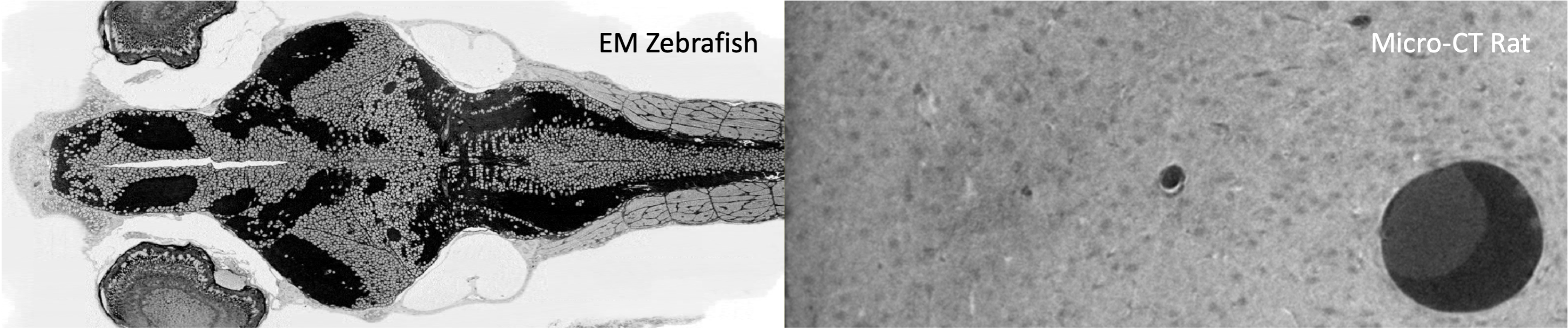
Segmenting 3D cell nuclei from microscopy image volumes is critical for biological and clinical analysis, enabling the study of cellular expression patterns and cell lineages. We pushed the task forward to the sub-cubic millimeter scale and curated the NucMM dataset with two fully annotated volumes: one 0.1 mm^3 electron microscopy (EM) volume containing nearly the entire zebrafish brain with around 170,000 nuclei; and one 0.25 mm^3 micro-CT (uCT) volume containing part of a mouse visual cortex with about 7,000 nuclei.
Please check our MICCAI 2021 paper for dataset details.

** Overview of the NucMM challenge dataset. NucMM contains two large volumes for 3D nuclei instance segmentation, including **(left) the NucMM-Z electron microscopy (EM) volume covering nearly a whole zebrafish brain, and (right) the NucMM-M micro-CT volume from the visual cortex of a mouse.
Dataset¶
[Data Release] Raw images and training/validation annotations can be downloaded here. If you want to access the complete annotation of the two NucMM volumes, please use this data request form.
 NucMM dataset characteristics.
NucMM dataset characteristics.
The annotation is not perfect. Please email linzudi@g.harvard.edu the (x,y,z) location of erroneous segmentation or the instance index to refine it together :) The subject of the email: "[NucMM Error]".
Citation¶
@inproceedings{lin2021nucmm, title={NucMM Dataset: 3D Neuronal Nuclei Instance Segmentation at Sub-Cubic Millimeter Scale}, author={Lin, Zudi and Wei, Donglai and Petkova, Mariela D and Wu, Yuelong and Ahmed, Zergham and Zou, Silin and Wendt, Nils and Boulanger-Weill, Jonathan and Wang, Xueying and Dhanyasi, Nagaraju and others}, booktitle={International Conference on Medical Image Computing and Computer-Assisted Intervention}, pages={164--174}, year={2021}, organization={Springer} }
Organizing committee¶
- Zudi Lin, School of Engineering and Applied Science, Harvard University, U.S.A.
- Donglai Wei, School of Engineering and Applied Science, Harvard University, U.S.A.
- Daniel Franco-Barranco (Donostia International Physics Center, Donostia-San Sebastian, Spain)
- Mariela D. Petkova, Molecular and Cellular Biology Department, Harvard University, U.S.A.
- Xueying Wang, Molecular and Cellular Biology Department, Harvard University, U.S.A.
- Ignacio Arganda-Carreras (Ikerbasque, Basque Foundation for Science, Bilbao, Spain)
- Florian Engert, Molecular and Cellular Biology Department, Harvard University, U.S.A.
- Jeff W. Lichtman, Jeremy R. Knowles Professor of Molecular and Cellular Biology, Santiago Ramón y Cajal Professor of Arts and Sciences, Harvard University, U.S.A.
- Hanspeter Pfister, An Wang Professor of Computer Science, School of Engineering and Applied Science, Harvard University, U.S.A.
Other Challenges¶
We have been organizing a series of connectomics segmentation challenges:
- MitoEM Challenge: Large-scale 3D Mitochondria Instance Segmentation
- AxonEM Challenge: 3D Axon Instance Segmentation of Brain Cortical Regions